More Information
Submitted: July 06, 2021 | Approved: July 19, 2021 | Published: July 20, 2021
How to cite this article: Luisetto M, Tarro G, Edbey K, Almukthar N, Cabianca L, et al. The SARS CoV-2 spike domain, RGD and integrin binding effect-relationship for vaccine design strategy. Ann Proteom Bioinform. 2021; 5: 027-041.
DOI: 10.29328/journal.apb.1001014
Copyright License: © 2021 Luisetto M, et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Keywords: SARS CoV-2; Spike domain; RGD; Integrin; Fibronectin; Protrombotic; Vaccine design strategy; Second generation COVID Vaccine
The SARS CoV-2 spike domain, RGD and integrin binding effect-relationship for vaccine design strategy
Luisetto M1*, Tarro G2, Khaled Edbey3, Almukthar N4, Cabianca L5, Mashori GR6, Yesvi AR7 and Latyschev OY8
1IMA, Academy Vice President, Applied Pharmacologist, Natural Science Branch, 29121 Italy
2President of the T & L de Beaumont Bonelli Foundation for Cancer Research, Naples, Italy
3Professor, Department of Chemistry, University of Benghazi, Libya
4Professor, Department of Physiology/College of Medicine, University of Babylon, Iraq
5Science and Technique Laboratory Degree Health City Hospital, Turin, Italy
6Professor, Department of Medical & Health Sciences for Woman, Peoples University of Medical and Health Sciences for Women, Pakistan
7Founder and President, Yugen Research Organization; Undergraduate Student, Western Michigan University, MI, 49008 USA
8IMA, Academy President, RU
*Address for Correspondence: Luisetto M, IMA, Academy Vice President, Applied Pharmacologist, Natural Science Branch, 29121 Italy, Email: [email protected]
Related the need to search new strategy in vaccine design in order to reduce also some rare effect like trombosys for some registered products it is interesting the role played by the SPIKE RGD domain.
The binding with molecules like Fibronectin is a process that must to be deeply investigated.
A better understanding in this process can be used to improve safety of the new generation of COVID vaccine.
The rare effect like thrombosis recognized by regulatory agency produced a modification of technical data sheet of some vaccine so the phenomena Is interesting to be more investigated.
Spike protein and its domains are involved in producing pathological effect of the COVID-19 disease.
What it is interesting is that some pathological effect of this pathology are similar to some rare side effect produced by some COVID-19 vaccine classes.
After a review of interesting literature related this topics is submitted an experimental projects able to verify in vitro the spike procoaugulant property.
Aim of this research study is to analyze under a biomolecular aspects some COVID-19 vaccine production strategy to produce knowledge useful to reduce if possible some rare side effect as reported by international drugs agency in last periods.
The COVID-19 Spike protein is a relevant molecule under a deeply investigation in the vaccine production by pharmaceutical industry.
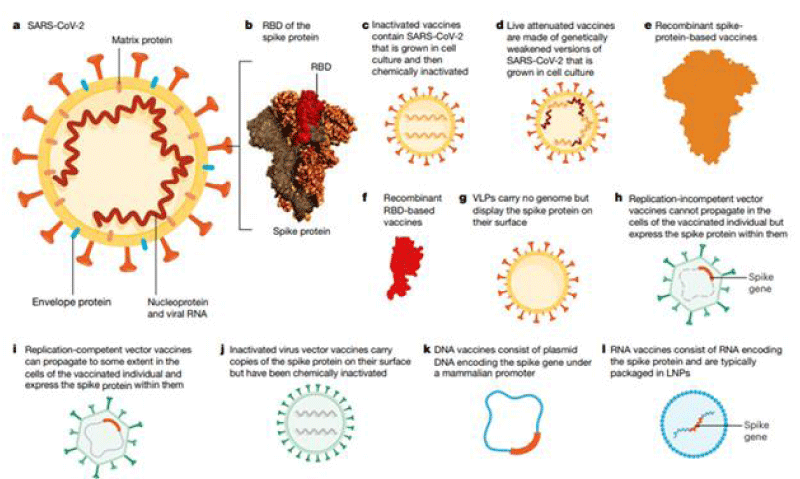
Different strategy are used: m-RNA vaccines, based on viral vector, based on spike protein and other (Figure 1).
Figure 1: The overview depending on which the scientists and researchers are targeting vaccine-developments.From: https://doi.org/10.1038/s41586-020-2798-2793.
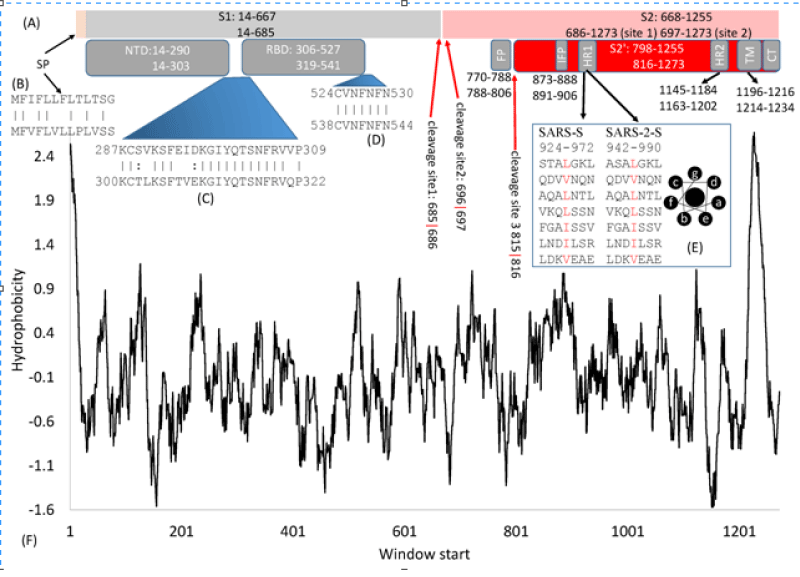
Some relevant characteristic of this molecule are crucial for the aim of this work: In article: Domains and Functions of Spike Protein in SARS-Cov-2 in the Context of Vaccine Design by Xuhua Xia, et al. written: The domain- structure is very similar between the SARS-S (UniProtKB: P59594) and the SARS-2-S (UniprotKB: P0DTC2). Both are cleaved to generate S1 and S2 subunits at specific cleavage -sites. S1 serves the function of receptor-binding and contains a signal- peptide (SP) at the N- terminus, an N-terminal domain (NTD), and receptor-binding- domain (RBD). S2 functions in membrane- fusion to facilitate cell -entry, and it contains the fusion peptide (FP) domain, internal fusion- peptide (IFP), 2 heptad-repeat domains (HR1 and HR2), transmembrane- domain, and a C-terminal domain (Figure 2).
Figure 2: Domain- structure of the SARS-S and the SARS-2-S. (A) Key -domains in SARS-S and SARS-2-S. SP, signal peptide; NTD, N-terminal domain; RBD, receptor-binding- domain; FP, fusion peptide; IFP, internal -fusion peptide; HR, heptad repeats; TM, transmembrane- domain; CT, cytoplasmic- tail. The top and bottom numbers in each domain pertain to SARS-S and SARS-2-S, in respectively way. The red- arrows indicate cleavage sites, and their numbers pertain to SARS-2-S; (B) Alignment of SP between SARS-S (top) and SARS-2-S ( see bottom); (C,D) Alignment of 2 inter-domain segments; (E) HR1 in SARS-S and SARS-2-S, together with the top view of a helix showing hydro-phobic- positions a and d on the same side; (F) Hydrophobicity plot generated from DAMBE.
Mohamed Hassan Kamel, et al: The pro-thrombotic- milieu in COVID-19 patients is generated by many factors of which stimulation of the extrinsic coagulation assumes a central -stage. Damage to the endothelial -cells activates TF which serves as the primary- trigger for the extrinsic coagulation- cascade.
Several components of Cytokine- storm such as tumor -necrosis factor alpha (TNF-α), interleukin-1 β (IL-1β) and interleukin-6 (IL-6) (through their cognate receptors and hypoxia associated with shock) upregulate TF mRNA in the Endothelial- cells.
Other cells involved in thrombotic- process such as platelets and polymorphonuclear cells (PMNs) are activated in the milieu of cytokine- storm and secrete TF-laden micro-particles that augment thrombo-genesis. SARS-CoV-2 RNA in the blood can serve as an activator of extrinsic coagulation- cascade. IL-6 augments thrombosis by several mechanisms. IL-6 increases the secretion of pro-thrombotic von Willerbrand facror (VWF) and factor VIII from liver and induces thrombocytosis in the bone- marrow. It also downregulates protein S in blood, a known inhibitor of thrombogenesis.
And according Liangwei Duan, et al: “SARS-CoV-2 S Protein Structure and Function: the SARS-CoV-2 S glycol-protein plays pivotal roles in viral -infection and in the covid-19 pathogenesis.
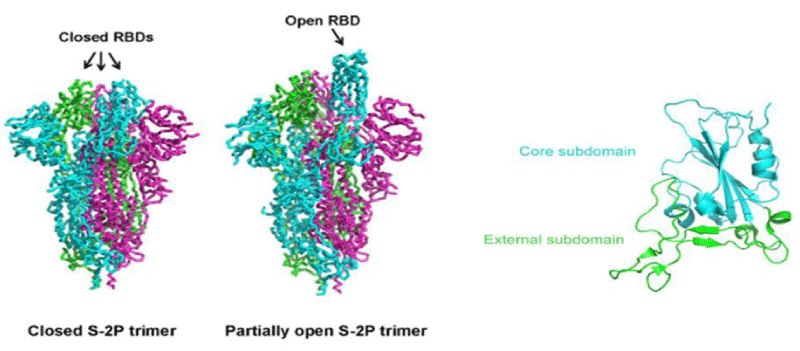
The overall -architecture of the prefusion SARS-CoV-2 S ectod-omain stabilized by 2 consecutive proline -mutations in 2 conformations determined by single particle cryo-electron microscopy (cryo-EM) is a ~160 Å long trimer with a triangular cross-section, with the S1- subunit adopting a “V” shape contributing to the overall triangular appearance and the S2- subunit forming the stalk. The structural difference between these 2 conformations only lies in the position of 1 of the three S1 RBDs. When all three RBDs are in the “down” position, the resulting S ectodomain- trimer assumes a closed- conformation, in which the receptor-binding surface of the S1 RBD is buried at the interface between protomers and cannot be accessible by its receptor. The S ectodomain- trimer with one single RBD in the “up” position assumes a partially open conformation and represents the functional- state, as the receptor-binding surface of the “up” RBD can be fully -exposed. The structural- information provides a blue-print for structure-based design of vaccine immunogens and entry inhibitors of the SARS-CoV-2.
Although the S -proteins of both SARS-CoV and SARS-CoV-2 are thought to be promising vaccine immunogens for generating protective- immunity, optimizing antigen design is critical to ensure an optimal immune- response through exposing more neutralizing- epitopes and displaying fewer potentially weakly or non-neutralizing epitopes. Vaccines containing or expressing full-length S protein or its soluble- ectodomain form should thus be engineered to sample a RBD(s) “up” conformation while the rest is still kept in the prefusion- state.
Although the SARS-CoV-2 S protein shares a high degree of amino -acid AA sequence identity with that of SARS-CoV (~76% overall), the RBM is less- conserved (~47% identity) than any other functional- region or domain, it can thus been surmised that the RBM has the most immune-dominant neutralizing epitope(s) of the whole S protein, capable of readily eliciting strong neutralizing antibody -responses.
Another concern for vaccine -candidates based on the full-length S glycoprotein of SARS-CoV-2 is raised by the observation that the S1- subunit could spontaneously dissociate from the S- glycoprotein probably as a trimer that still assumes the RBD closed -conformation, leaving only the post-fusion S2 trimmer. The resulting S1 and S2 sub-units might expose immune-dominant, non-neutralizing epitopes that are utilized by SARS-CoV-2 to serve as decoys to distract the host- immune system, inducing a large proportion of ineffective antibody -responses, as documented for HIV-1 and respiratory syncytial -virus (RSV).
Even if vaccine- candidates based on the full-length S- protein of the closely related SARS-CoV could elicit neutralizing antibody- responses against infection of SARS-CoV, they may also induce harmful- immune responses, including liver- damage of the vaccinated animals, infection of human immune- cells by SARS-CoV, and antibody-dependent enhancement of SARS-CoV infection. Although the S proteins of both SARS-CoV and SARS-CoV-2 are thought to be promising vaccine- immunogens for generating protective- immunity, optimizing antigen design is critical to ensure an optimal immune- response through exposing more neutralizing epitopes and displaying fewer potentially weakly or non-neutralizing epitopes. Vaccines containing or expressing the full-length S- protein or its soluble ecto-domain form should thus be engineered to sample a RBD(s) “up” conformation while the rest is still kept in the prefusion- state.
Apart from recombinant, soluble, stabilized- ectodomains that are engineered to expose the immunodominant RBD by adapting the RBD(s) “up” conformation, RBD- proteins of SARS-CoV and SARS-CoV-2 have also been widely used as recombinant protein-based vaccines. The RBD of the SARS-CoV is highly immunogenic and is targeted by most of the neutralizing monoclonal- antibodies MABS that have been characterized. Based on the observation that a 193-amino acid- fragment (residues 318-510) was previously identified to be the minimal RBD- region of SARS-CoV, a corresponding 194-amino acid- fragment (residues 331-524) can be readily selected as the minimal RBD -region of SARS-CoV-2 and has already been characterized. This minimal form of RBDs of both viruses could serve as a vaccine candidate.
The epitope-focused vaccine design has proven to be successful in generating neutralizing -antibodies against RSV fusion-glycoprotein. Wide-type full-length or soluble ecto-domain form of the SARS-CoV-2 S protein could trigger stronger cellular- immune responses (Figure 3).
Figure 3: The different situation and trimer-based RBD in a glimpse. From: https://doi.org/10.1016/j.fmre.2021.01.009.
Jonaid Ahmad Malik, et al: “The potential components of S -protein for use as antigens include the FLSP, the RBD- domain, the S1 subunit, the S2- subunit, the N-terminal domain (NTD), and membrane fusion- peptide (FP).
BioNTech/Fosun Pharma/Pfizer on the other hand are working on a unique combination of m-RNA- sequences as a vaccine candidate which contains 2 nucleoside modified m-RNA candidates (BNT162a1 andBNT162b2). BNT162a1 is a uridine containing m-RNA candidate whileBNT162b2 is a m-RNA.
BNT162b1 consists of SARS-CoV-2 receptor binding- domain (S2RBD), while BNT162b2 contains the SARS-CoV-2FLSP antigen. All of these m-RNA constructs are packed within an LNP-preparation.
The spike protein is incorporated in 2 of the candidates and a smaller RBD fragment of the spike protein is incorporated in the other 2 candidates. On 9th November 2020, Pfizer and BioNTech announced that their vaccine candidate demonstrated evidence of efficacy in 90 % of participants, who were not exposed to SARS-CoV-2infection before injection, based on the first interim efficacy- analysis from the Phase 3 clinical- study.
The investigation further affirmed SARS-CoV RBD (S-RBD) generated antibodies that cross-react with S2-RBD protein, and S-RBD-actuated antisera cross-kill SARS-CoV-2, recommending the possibility to create SARS-CoV RBD-based immunizations for counteraction of the SARS-CoV2 infection.
Most advanced spike protein-based candidate, ChAdOx 1 vaccine developed by the University of Oxford Jenner -Institute. The vaccine contains the optimized full-length surface S- glycoprotein sequence of SARS-CoV-2, with a TPA as a leader- sequence. It uses a RDCA to deliver aSARS-CoV-2 S protein to induce a protective immune- response. The evidence for the effectiveness of the ChAdOx 1 was increased after researchers confirmed that the single-dose protected 6 rhesus –macaques from the incidence of pneumonia invoked by SAR-COV-2 (Figure 4).
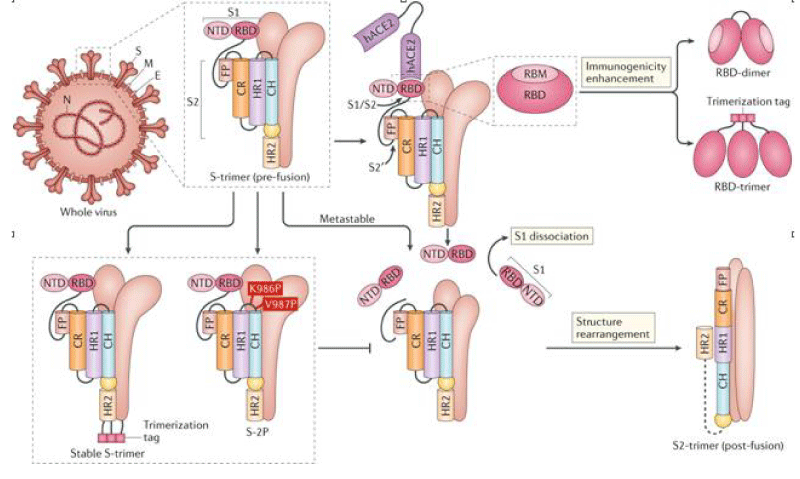
Figure 4: SARS-CoV-2)contains 4 major structure proteins: spike (S), membrane (M) and envelope (E) proteins, which are embedded on the virion -surface, and nucleocapsid (N) protein, which binds viral -RNA inside the virion. The S- protein trimer in its pre-fusion conformation is shown. The S- protein comprises the S1 subunit (which includes the N-terminal domain (NTD) and the receptor-binding domain (RBD)) (the receptor-binding motif (RBM) within the RBD is also labelled) and the S2 subunit (which includes fusion peptide (FP), connecting- region (CR), heptad repeat 1 (HR1), heptad repeat (HR2) and central- helix (CH)). The SARS-CoV-2 S protein binds to its host- receptor, the dimeric human angiotensin-converting enzyme 2 (hACE2), via the RBD and dissociates the S1- subunits. Cleavage at both S1–S2 and S2′ sites allows structural re-arrangement of the S2 subunit required for virus–host membrane fusion. The S2-trimer in its post-fusion arrangement is shown. The RBD is an attractive vaccine- target. The generation of an RBD-dimer or RBD-trimer has been shown to enhance the immune-genicity of RBD-based vaccines. A stabilized S-trimer shown with a C-terminal trimer-tag is a vaccine target. The pre-fusion S protein is generally metastable during in vitro preparations and prone to transform into its post-fusion conformation. Mutation of 2 residues (K986 and V987) to proline stabilizes S- protein (S-2P) and prevents the pre-fusion to post-fusion structural- change.
Lianpan Dai, et al: Learning from the previous experience with these CoVs, the S-2P design is now being used in several vaccine- strategies against the COVID-19. SARS-CoV-2 S-2P (comprising proline- substitutions at residues K986 and V987) is used as the target antigen in three gene-based vaccine candidates (mRNA vaccines by Moderna/National Institute of Allergy and Infectious -Diseases (NIAID) and BioNTech/Pfizer and a recombinant Ad26 vaccine by Janssen Pharmaceutical -Companies) and a protein-based candidate (by Novavax). Mutation at the cleavage sites in the S- protein is also believed to stabilize the pre-fusion conformation of the S- protein. S-2P in the Janssen Ad26-vectored -vaccine (Ad26.COV2.S) and in the Novavax protein-based vaccine (NVX-CoV2373) contains additional mutations at the S1–S2 polybasic cleavage site from RRAR to SRAG or QQAQ to render it protease resistant, which helps to further stabilize the S- protein in its pre-fusion conformation.
The use of RBD in vaccines is compromised by its limited -immunogenicity owing to its small molecular- size and possible mixed forms of multiple- complexes (monomers, dimers or trimers). Strategies to overcome these drawbacks include increasing antigen -size (by fusing the RBD with an Fc domain) or by RBD- multimerization (by displaying multiple -copies of RBD on particles). To address these limitations, our teamwork described a generalizable strategy to design a dimeric form of the RBD of beta-CoV antigens suitable for use against SARS-CoV-2, MERS-CoV and SARS-CoV120. The RBDs from SARS-CoV, MERS-CoV or SARS-CoV-2 spontaneously form dimers in solution. Structural -analyses showed, for MERS-CoV118 and SARS-CoV121, that both RBD- protomers in a dimer stack on top of each other via the core -sub-domains and expose the RBM, the major site recognized by nAbs, indicating similar RBD-dimer structures for other CoVs. Structure-guided design yielded homogeneous RBD-dimers as a tandem-repeat single chain. The RBD-dimer- antigen induced 10-fold to 100-fold higher nAb titres than the conventional RBD-monomer and was protective in a mouse -model. We developed a COVID-19 vaccine candidate, ZF2001, comprising the RBD-dimer as the target. This protein subunit- vaccine is currently being evaluated in phase- III clinical -trials. In addition to the RBD-dimer, an mRNA vaccine, BNT162b1 (BioNTech/Pfizer), was reported to express an RBD-trimer stabilized by the fold on trimerization-domain.
S1-NTD and the S2 subunit
The S1-NTD contains epitopes for CoVn Abs found in infected- patients and has been considered a potential target in CoV -vaccines. NTD-targeting nAbs generally do not directly -block receptor binding but rather interfere with receptor- binding or restrain the S- protein conformational changes required for the pre-fusion to post-fusion transition. SARS-CoV-2 NTD-targeting nAbs generally exhibit lower- neutralizing potency than RBD-specific nAbs. We previously reported an NTD-based vaccine against MERS-CoV. Vaccination with NTD- protein elicited nAbs and NTD-specific T -cell responses. it reduced lung ab-normalities in a MERS-CoV challenge mouse -model, although the immune-genicity and protective efficacy of the NTD- protein were weaker than the RBD protein. The inclusion of NTD in a COVID-19 vaccine would broaden the neutralizing- epitopes and reduce the potential of viral- escape of host immunity. Yet, so far, NTD-based vaccines against COVID-19 have not been reported
For the S2- subunit, peptides derived from HR1 or HR2 of the S2- subunit from SARS-CoV, MERS-CoV and SARS-CoV-2 have been described that inhibit viral- fusion with target cells and thereby prevent virus infection. nAbs have been reported to target the S2- sub-unit of CoVs, including SARS-CoV-2, suggesting the S2 subunit as a COVID-19 vaccine target.
The membrane-proximal S2- subunit contains more extensive N-glycan shielding and is less accessible for immune recognition than the S1 -subunit and is therefore less- immunogenic. Rabbits immunized with SARS-CoV-2 S2 protein showed much lower nAb- titres than those immunized with the S1 -subunit or RBD proteins. S2 subunit-targeting antibodies isolated from convalescent patients showed weaker neutralizing- activities against SARS-CoV-2 than RBD-targeting antibodies. These studies suggest that the S2- subunit alone may not be an effective -target for humoral- responses. Because of the relative sequence conservation of the S2 -subunit between virus- species, the S2 subunit is targeted by cross-reactive antibodies and CD4+ T cells recognizing both SARS-CoV-2 and other human CoVs, suggesting a potential target in universal CoV- vaccines.
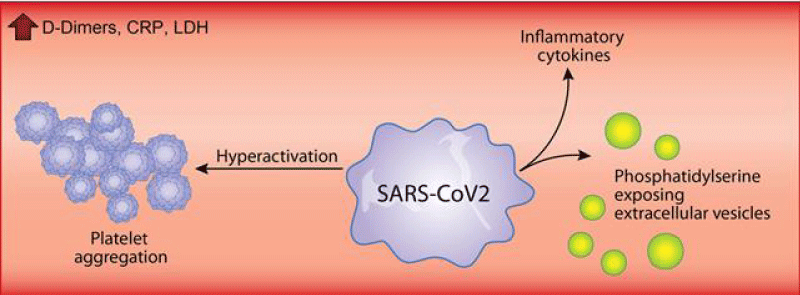
Younes Zaid, et al: In addition to the over whelming lung -inflammation that prevails in coronavirus -disease 2019, hyper-coagulation and thrombosis contribute to the lethality of subjects infected with severe acute respiratory -syndrome coronavirus- 2. Platelets are chiefly implicated in thrombosis. They can interact with viruses and are an important source of inflammatory- mediators. While a lower- platelet count is associated with severity and mortality, little is known about platelet -function during the COVID-19 (Figure 5).
Figure 5: Sars cov-2 effect.
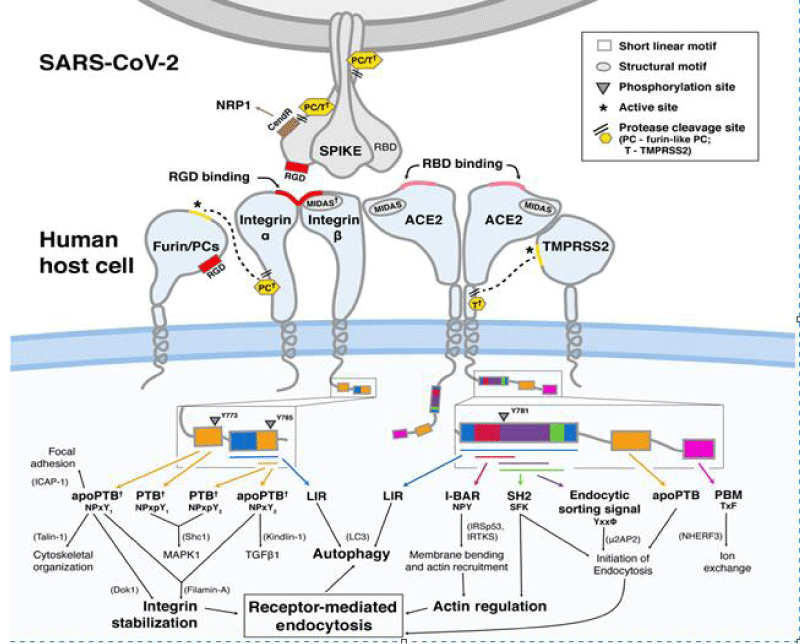
So it is interesting to observe the initial link between coronavirus and the epithelia lung cells and the biologic molecular implication: especially for the RGD and RBD domain interaction (Figures 6-8).
Figure 6:
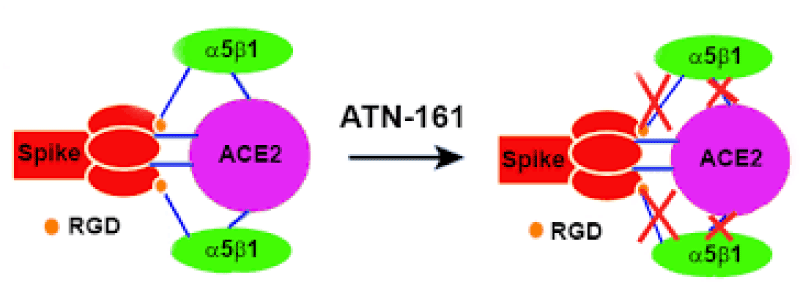
Figure 7: The Integrin Binding Peptide, ATN-161, as a Novel Therapy for SARS-CoV-2 Infection. Brandon Beddingfield, et al.
Figure 8: Another secret of SARS-COV-2 revealed!
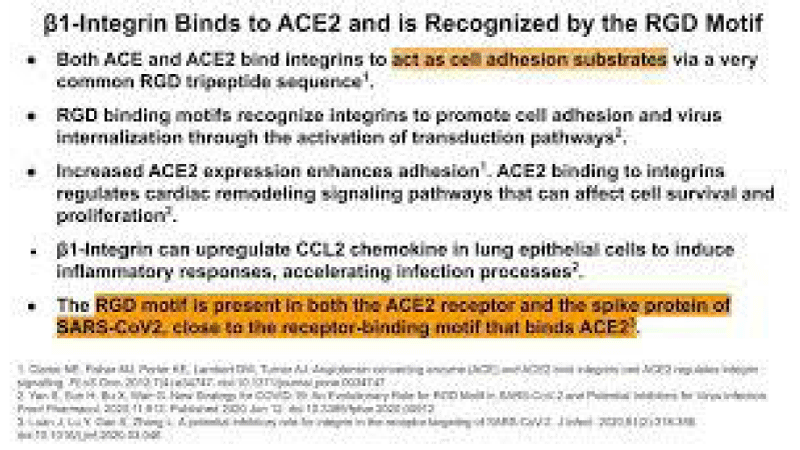
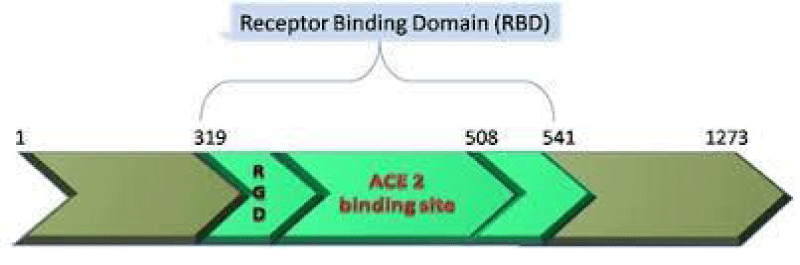
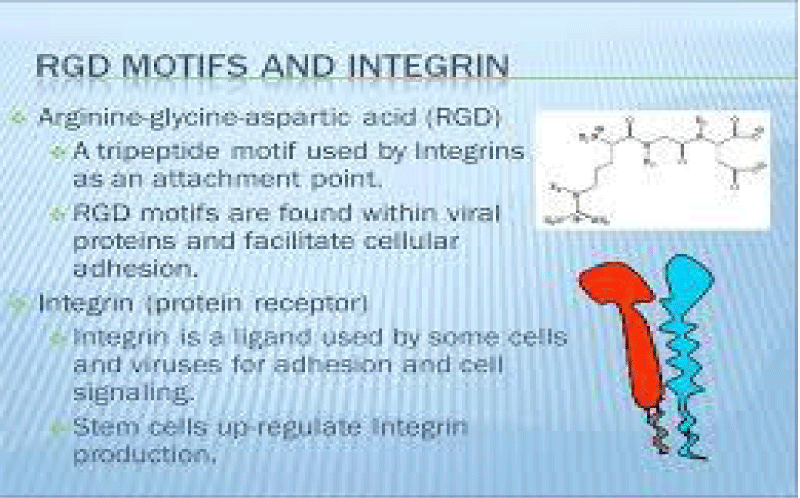
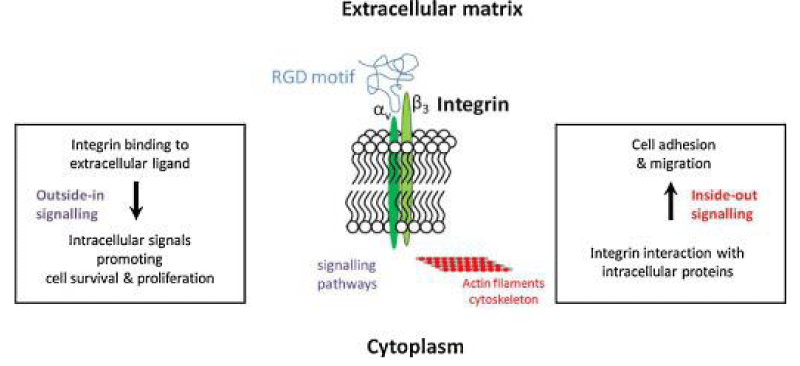
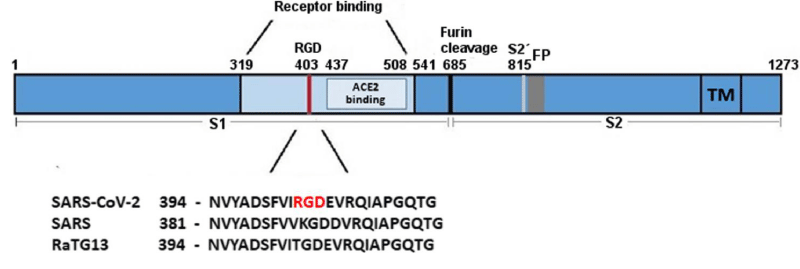
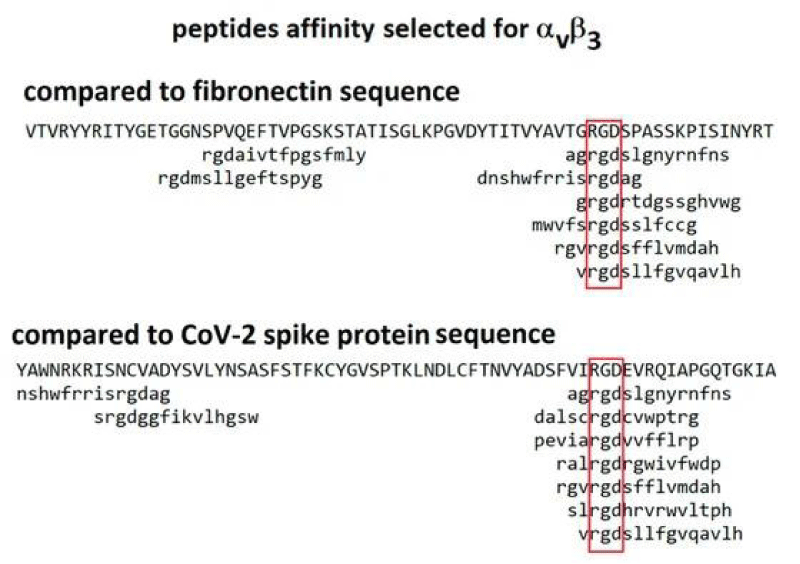
P Singh Kshatriya: A very well-known conserved RGD (403–405: Arg-Gly-Asp) - motif is located few amino acids AA upstream of the ACE2- binding- region in the binding -domain of spike protein of the SARS-CoV-2. This motif is absent in all the other corona-viruses known till date. This RGD motif is crucial and adequate enough for the virus-human- cell binding in various other human -viruses such as HMPV, HHV, RV, etc. This motif is present in antigen of the viruses allows it to utilize the integrins present on the host- cells as its receptors. This is essential to form the first level of contact for its viral- entry into the cells.
This finding reveals integrin- binding of the virus as a promising therapeutic- target. Genaxxon’s SARS-CoV-2 (2019-nCoV) Spike S1- Protein (RBD) peptide would be an ideal- peptide to work with in this direction. Genaxxon also has a wide -range of viral- peptides and are worth a try > RGD binding- motifs are extremely important peptide in targeted- therapy and included in various bio-materials designed to promote wound healing. This motif in viruses has been shown to enhance their patho-genicity by activating various signaling- pathways like MAPK and the PI-3K. It has been pointed out that binding to integrin may play a supplemental role to ACE2 binding, like facilitating endocytosis by signaling through the integrin. The virus could infect different target- cells by binding to ACE2 or to integrins. Blockers of RGD-binding integrin could be another way of tackling SARS-CoV-2 (Figures 9,10).
Figure 9: Bálint Mészáros, et al. Short linear- motif candidates in the cell entry system used by SARS-CoV-2 and their potential therapeutic implications. Research Resource biochemistry.
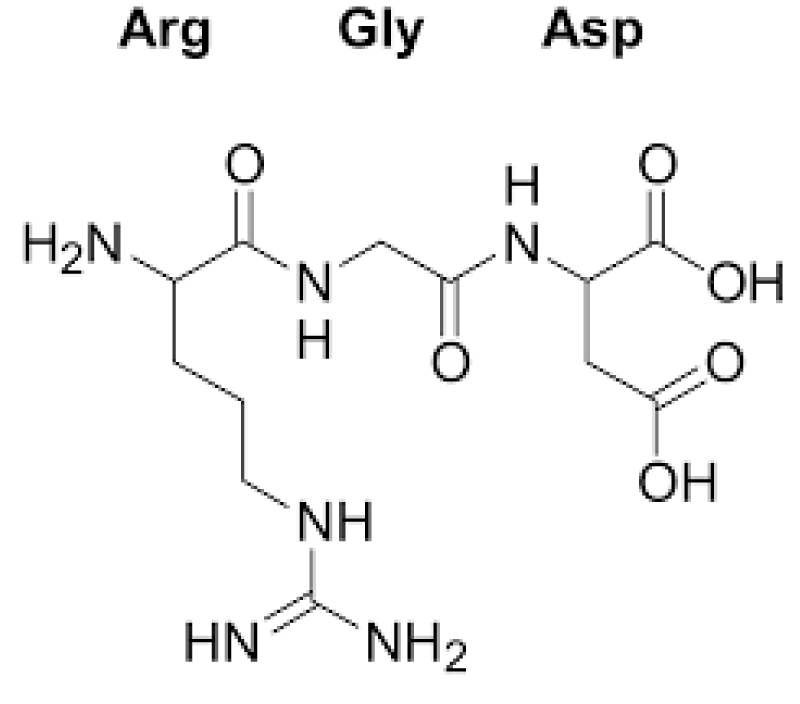
Figure 10: RGD.
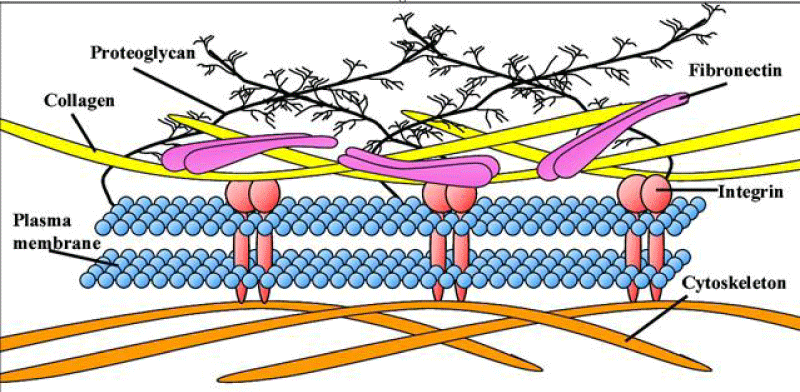
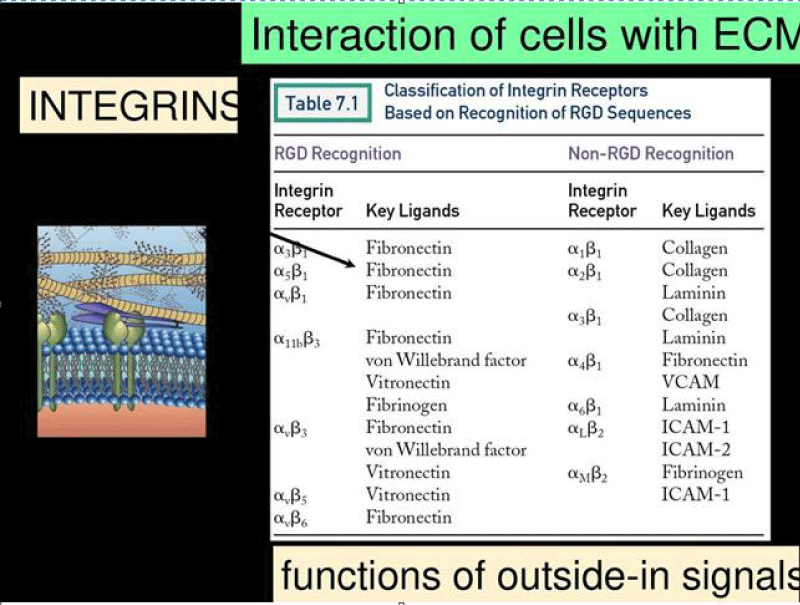
The tri-peptide Arg-Gly-Asp (RGD) consists of Arginine, Glycine, and Aspartate AA. It was identified as the amino acid- sequence within the extracellular matrix protein fibronectin that mediates cell- attachment. The RGD cell binding sequence has since been identified in other extracellular- matrix proteins, including vitronectin and the laminin. The family of membrane- proteins known as integrins act as receptors for these cell adhesion -molecules via the RGD- motif. A subset of the integrins recognize the RGD- motif within their ligands, the binding of which mediates both cell-substratum and cell-cell interactions. These integrins include αvβ3, α5β1 and αIIbβ3.
The RGD- domain is both sufficient - indispensable for cell- membrane binding. As such, the RGD cell binding motif has relevance in the fields of oncology, tissue- engineering and in the regenerative -medicine. Because of their cell-adhesive activity, RGD- peptides are frequently incorporated into bio-materials designed to promote wound healing. RGD is an important peptide sequence often used in the Targeted -therapy. The RGD peptide can be used to target cancer cells on which cell membrane integrins are up-regulated compared to healthy -cells.
Though the minimum sequence peptide RGD retains the property of cell- adhesion, integrin-RGD bonds are consider-ably weaker than integrin-fibronectin bonds. The reduced adhesion strength is likely due to both lack of secondary structure and other synergistic domains, namely PHSRN, present in the full-length protein Full- length protein domains differentially mediate cell morphology, cell- migration and cell proliferation compared to RGD alone.
Other minimum sequence- motifs have been identified, including the GFOGER 30 amino- acid AA sequence from collagen and the YGISR and A5G81sequences from laminin.
Fibronectin is a high-molecular weight (~500 kDa) glycol-protein of the extra-cellular- matrix that binds to membrane-spanning receptor proteins named integrins. Fibronectin binds to other extra-cellular matrix proteins like: collagen, fibrin, heparan sulfate -proteoglycans (the syndecans).Fibronectin has various functions that ensure the normal -functioning of vertebrate. It is clear involved in cell- adhesion, growth, migration, and in the differentiation-process. Cellular- fibronectin is assembled into the extra-cellular- matrix, an insoluble- network that separates and supports the organs and tissues of an organism.
Fibronectin plays a crucial role also in the in wound- healing. Along with fibrin, plasma fibronectin is deposited at the site of the injury, forming a blood- clot that stops bleeding and protects the underlying tissue. As repair of the injured- tissue continues, fibro-blasts and the macrophages begin to remodel the area, degrading the proteins that form the provisional blood- clot matrix and replacing them with a matrix that more resembles the normal, surrounding- tissue.
Figure 11:
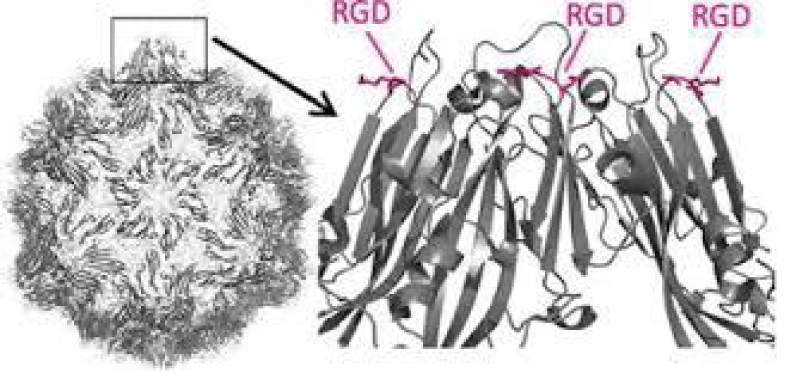
Figure 12: Xingjie Zana, et al. Polyvalent display of RGD motifs on turnip yellow mosaic virus for enhanced stem- cell adhesion and spreading.
Many functional and protein-binding domains along the length of a fibronectin -monomer. There are 4 fibronectin-binding domains, allowing fibronectin to associate with other fibronectin -molecules. One of these fibronectin-binding domains, I1–5, is referred to as the "assembly- domain", and it is required for the initiation of fibronectin -matrix assembly. Modules III9–10 correspond to the "cell-binding -domain" of the fibronectin. The RGD- sequence (Arg–Gly–Asp) is located in III10 and is the site of cell- attachment via α5β1 and αVβ3 integrins on cell- surface. The "synergy- site" is in III9 and has a role in modulating fibronectin's association with α5β1- integrins. Fibronectin contains domains for fibrin-binding (I1–5, I10–12), collagen-binding (I6–9), fibulin-1-binding (III13–14), heparin-binding and syndecan-binding (III12–14).
Yiming Wang, et al: Fibronectin (Fn) is an essential extra-cellular- matrix protein involved in cell -adhesion, migration, differentiation and in proliferation. Fn is required for embryogenesis, and is intricately involved in malignant transformation, angiogenesis, inflammation, fibrosis, wound healing, thrombosis and hemostasis. Fn is a dimer consisting of two 250 kDa subunits. Alternative splicing of Fn pre-mRNA at extra domain (ED)-A, ED-B, and variable -region results in up to 20 Fn variants in humans, which are categorized into 2 major groups, cellular- fibronectin (cFn) and plasma fibronectin (pFn). cFn contains at least one of the EDs and is expressed by various cell- types. In contrast, pFn is secreted specifically by hepatocytes to the blood- circulation and excludes both ED-A and ED-B. Since pFn is an abundant protein in the blood- plasma and contains integrin -binding motif Arg-Gly-Asp (RGD) as well as N-terminal fibrin and collagen binding- sites, it has long been suspected to play a role in thrombosis and in hemostasis process. Our previous study using pFn-/- mice showed that pFn promotes thrombus growth and stability in vivo.
Fibronectin is an extra-cellular multidomain glycoprotein that directs and regulates a variety of cell- processes such as proliferation, development, haemostasis, embryogenesis, and wound- healing. As a major component of blood, fibronectin exists as a soluble di-sulphide linked- dimer, but it can also be incorporated into an insoluble cross-linked fibrillar network to form a major component of the extra-cellular- matrix. Fibronectin is composed of an extended -chain of module repeats termed Fn1, Fn2, and Fn3 that bind to a wide- range of transmembrane receptors and extracellular matrix components, including the collagen.
Platelets and platelet allo-antigens: Lessons from human patients and animal models of fetal and neonatal allo-immune thrombocytopenia. Vadasz B, et al. (Figures 13-17).
Figure 13: Structural and functional characterisation of the collagen binding domain of fibronectin. CJ Millard 2007.
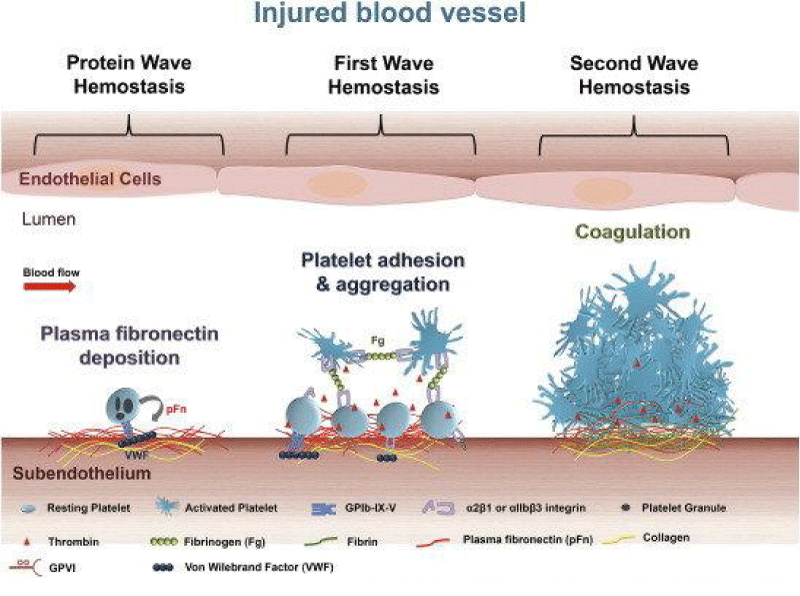
Figure 14: Roles of platelets in thrombosis - hemostasis. After vascular injury, plasma- fibronectin quickly deposits onto the injured vessel- wall. Platelets may also release their internalized plasma -fibronectin from their granules. These plasma and platelet sources of fibronectin likely synergistically contribute to the protein- wave of hemostasis. Platelet adhesion - aggregation ( the classical first wave of hemostasis) are then initiated via platelet- receptors and their ligands. Activated -platelets also provide a negatively -charge surface and mediate cell-based thrombin generation, which contributes to blood coagulation that is initiated following tissue- damage ( the second wave of hemostasis). In a growing hemostatic plug/thrombus, the fibrin and fibronectin- matrix is usually formed in the interface between the injured vessel wall and platelet- plug.
Figure 15: Eustacia McKenzie.
Figure 16: N Withofs, R Hustinx. Integrin αvβ3 and RGD-based radiopharmaceuticals L’intégrine αvβ3 et l’imagerie par les radio-pharmaceutiques de type RGD. https://doi.org/10.1016/j.mednuc.2015.12.002.
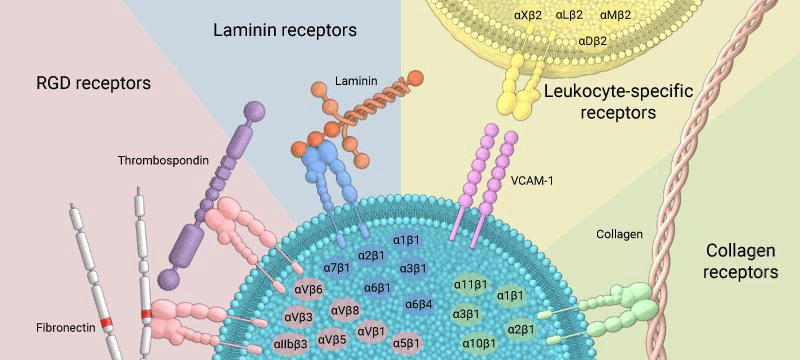
Figure 17: Integrins can bind to a diverse range of ligands, which can be broadly categorized into: RGD receptors, laminin receptors, leukocyte-specific receptors, and collagen receptors.
Common ECM -components that are bound by integrins (with respective recognition- sequence) are:
Fibronectin (RGD, LDV)
Collagen (triple helical GFOGER)
Laminin
(RGD)
Fibrinogen (RGD)
Thrombospondin
Glycoproteins (e.g. tenascin C, osteopontin, nefronectin).
Immunologically important integrin- ligands are the inter-cellular adhesion- molecules (ICAMs), immunoglobulin IG superfamily members present on inflamed endothelium and antigen-presenting cells.
Integrins are broadly grouped into 4 categories based on their ligand-specificity:
RGD receptors (α5β1, αVβ3, αVβ1,αVβ5, αVβ6, αVβ8, and αIIbβ3).
Laminin receptors (α1β1, α2β1, α3β1, α6β1, α7β1, and α6β4).
Leukocyte-specific receptors (αLβ2, αMβ2, αXβ2, and αDβ2).
Collagen receptors (α1β1, α2β1, α3β1, α10β1, and α11β1).
With an observational method some relevant biomedical literature (for the scope of this work) is reported and analysed.
All literature comes from Pub med.
An expertimental hypothesis project is submitted in order to produce a global conclusion.
According Ana Shulla, et al: Enveloped viruses enter cells by viral glycoprotein-mediated binding to host- cells and subsequent fusion of virus and host cell- membranes. For the corona-viruses, viral spike (S)- proteins execute these cell entry functions. The S- proteins are set apart from other viral and cellular- membrane fusion proteins by their extensively palmitoylated membrane-associated tails.
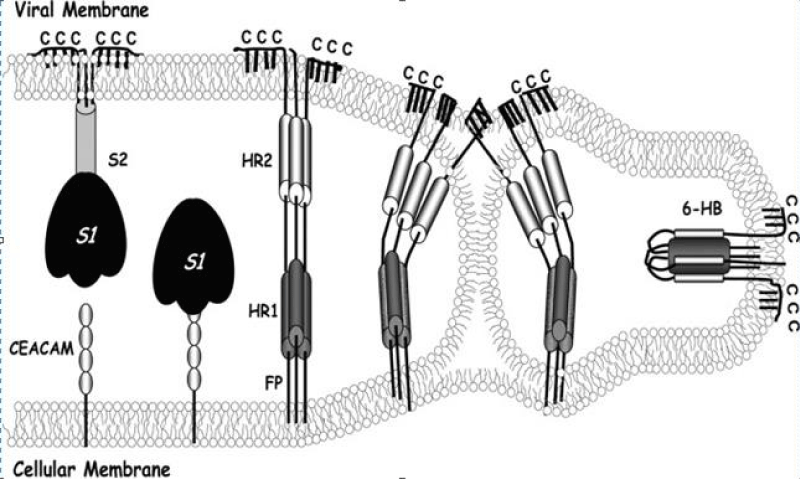
Biological- membranes are configured in large part by protein-mediated fission and fusion reactions. Enveloped- viruses can reveal the principles of these processes because their assembly and budding from infected -cells requires membrane fissions, and their entry into susceptible cells depends on membrane- fusions. Glycoproteins extending from virion- surfaces mediate the fusion process. These specialized integral membrane -proteins are in metastable high energy configurations on virus surfaces, and they drive coalescence of opposing virus and cell -membranes by undergoing a series of energy-releasing un-folding and re-folding events [1] (Figure 18).
Figure 18: Coronavirus S protein-mediated membrane fusion- model. A hypothetical depiction of the native prefusion S- protein (left) is depicted binding to cellular CEACAM- receptors. S1 subunit dissociation and S2 unfolding generates pre hairpin- structures (middle) depicted with cell membrane-intercalated FPs and exposed heptad repeat regions (HR1 and -2). Prehairpin closure through a lipid stalk intermediate generates a highly -stable, rodlike 6-HB, in which HR2- helices are positioned antiparallel to an interior HR1 trimer. Palmitates are shown extending from endo-domain- cysteines (C).
Severe acute respiratory syndrome coronavirus 2 (the SARS-Cov-2)-induced infection, the cause of coronavirus -disease 2019, is characterized by unprecedented clinical -pathologies. One of the most important pathologies, is hyper-coagulation and micro-clots in the lungs of patients. Here we study the effect of isolated SARS-CoV-2 spike protein S1- subunit as potential inflammagen sui generis. Using scanning electron and fluorescence- microscopy as well as mass- spectrometry, we investigate the potential of this inflammagen to interact with platelets and fibrin(ogen) directly to cause blood -hyper-coagulation. Using platelet poor plasma (PPP), we show that spike- protein may interfere with blood flow. Mass- spectrometry also showed that when spike protein S1 is added to healthy PPP, it results in structural changes to β and γ fibrin (ogen), complement, and prothrombin. These proteins were substantially resistant to trypsinization, in the presence of spike protein -S1. Here we suggest that, in part, the presence of spike -protein in circulation may contribute to the hyper-coagulation in COVID-19 positive patients and may cause substantial impairment of fibrinolysis. Such lytic impairment may result in the persistent large-micro-clots we have noted here and previously in plasma samples of COVID-19 patients. This observation may have important clinical -relevance in the treatment of hyper-coagulability in COVID-19 patients [2].
Vallabh Janardhan, et al: “Mechanisms of blood- clots and stroke in COVID-19: In Panel D, a cascade of events unfolds resulting in blood -clots and strokes. In Step 1, endothelial- dysfunction can occur either directly by viral entry into the endo-thelial cell or indirectly with the accumulation of Angiotensin- (Ang) II as SARS-CoV-2 attaches to ACE2 and is not able to convert Ang II to Ang 1,7. Ang II can result in reactive- oxygen species (ROS), oxidative stress, and endothelial- dysfunction. In Step 2, oxidation of beta 2 glycoprotein 1 (β2GP1) occurs due to endo-thelial dysfunction and ROS, and results in the formation of anti-phospholipid (aPL) antibody complexes. In Step 3, platelet- adhesion occurs and platelets attach to the sub-endothelial collagen using von Willebrand -Factor (vWF). This happens because the non-oxidized β2GP1 is no longer available to competitively bind vWF. In Step 4, platelet activation occurs due to platelets- binding to vWF resulting in granule-release (α and dense) and the presence of aPL complexes further promotes platelet -activation. In Steps 5-7, platelet aggregation using Vwf or fibrinogen (released by α granules), formation of thrombus via the coagulation -cascade, and subsequent cross-linking of fibrin strands to stabilize the clot occur. In Step 8, a pulmonary- embolism (PE) and/or a large vessel occlusion stroke occurs. Therapeutic implications:
In Panel E, several blood thinners like antiplatelet- drugs can impact Steps 4 and 5 (oral aspirin, oral clopidogrel), anti-thrombotic drugs can impact Step 6 (antithrombin III binding- agent, parenteral heparin, vitamin K- antagonist,, warfarin, newer direct Xa or thrombin- inhibitors), and fibrinolytic -drugs can impact Step 7 (intravenous tissue plasminogen- activator or tenecteplase). Several catheter-based devices (approved in EU, and cleared to market in the USA) can impact Step 8 (clot retrieval devices that try to use clot- integration, continuous vacuum aspiration pumps, and devices that try to use uniform negative suction pressure to ingest clots) or newer devices (approved in Eu) can impact Step 8 (cyclical vacuum- aspiration- pumps and devices that use pulsating negative -suction pressure to improve complete clot ingestion and reduce clot- fragmentation), help remove blood clots, and treat PE and large vessel- strokes [3].
Although ACE2 is considered the primary receptor for CoV-2 cell- entry, recent reports suggest that alternative pathways may contribute.
This article considers the hypothesis that viral binding to cell-surface integrins may contribute to the high infectivity and widespread extra-pulmonary impacts of the SARS-CoV-2 virus. This potential is suggested on the basis of the emergence of an RGD- (arginine-glycine-aspartate) sequence in the receptor-binding domain of the spike protein. RGD is a motif commonly used by viruses to bind cell-surface integrins. Numerous signaling- pathways are mediated by integrins and virion binding could lead to dysregulation of these path-ways, with consequent tissue -damage. Integrins on the surfaces of pneumo-cytes, endothelial cells and platelets may be vulnerable to CoV-2 virion binding.
Binding of intact -virions to integrins on alveolar- cells could enhance viral -entry. Binding of virions to integrins on endothelial- cells could activate angiogenic- cell signaling- pathways; dysregulate integrin-mediated signaling pathways controlling developmental -processes; and precipitate endothelial-activation to initiate blood clotting. Such a pro-coagulant state, perhaps together with enhancement of platelet- aggregation through virions binding to integrins on platelets, could amplify the production of micro-thrombi that pose the threat of pulmonary thrombosis and embolism, strokes and other thrombotic- consequences. The susceptibility of different tissues to virion–integrin interactions may be modulated by a host of factors, including the conformation of relevant integrins and the impact of the tissue micro-environment on spike- protein conformation. Patient-specific differences in these factors may contribute to the high- variability of clinical presentation. There is danger that the emergence of receptor-binding domain mutations that increase infectivity may also enhance access of the RGD motif for integrin- binding, resulting in viral- strains with ACE2 independent routes of cell entry and novel integrin-mediated biological and clinical- impacts. The highly infectious variant, B.1.1.7 (or VUI 202012/01), includes a receptor-binding domain amino acid AA replacement, N501Y, that could potentially provide the RGD- motif with enhanced access to cell-surface integrins, with consequent clinical- impacts” [4] (Fifure 19).
Figure 19:
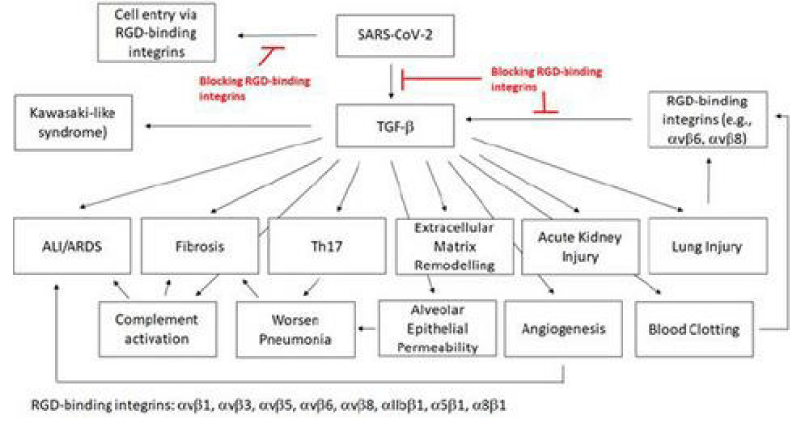
SARS‐CoV‐2 is a global pandemic and a severe public health -crisis. SARS‐CoV‐2 is highly contagious and shows high -mortality rates, especially in elderly and patients with pre‐existing medical -conditions. At the current stage, no effective drugs are available to treat these patients. In this work, we analyse the rationale of targeting RGD‐binding integrins to potentially inhibit viral- cell infection and to block the TGF‐β activation, which is involved in the severity of several human- pathologies, including the complications of the severe COVID‐19 cases.
We demonstrate the correlation between the ACE2 and TGF‐β expression and the possible consequences for the severe COVID‐19 infections. we list approved drugs or drugs in clinical- trials for other diseases that also target the RGD‐binding integrins or TGF‐β. These drugs have already shown a good safety -profile and, therefore, can be faster brought into a trial to treat COVID‐19 patients [5].
The findings of blood -clots in COVID‐19 patients and their involvement from deep venous- thrombosis in lower extremities to blocked arteries in the brain and lungs, resulting in strokes and pulmonary- embolism, are of great concern and interest. The source of these blood -clots has not been defined; TGF‐β is known to stimulate the production of Factor XII (FXII). The zymogen FXII stands at the beginning of the coagulation -signalling of the intrinsic- cascade Thrombin, which is the last step of the coagulation- cascade, mediates the cleavage of fibrinogen to fibrin. The fibrin- monomers polymerise and form a fibrin -clot that stop the bleeding. FXII knock-out mice are largely defective for thrombus formation and are protected from experimental cerebral ischaemia and pulmonary -embolism. FXII, thrombin and fibrinogen have also been linked to inflammatory disorders like multiple sclerosis, sepsis, rheumatoid -arthritis and colitis. Thrombin cleaves GARP (glycoprotein- A repetitions predominant) that is found on the surface of platelets, and the cleaved GARP in cooperation with αvβ8 leads to the release of mature TGF‐β1, creating a positive feedback- loop. Blocking αvβ8 on dendritic -cells and fibroblasts impairs TGF‐β‐dependent generation of Th17 -cells and reduces the inflammatory -disease in the lungs. It has been postulated that FXII potentially mediates the development of neuro-inflammation via upregulation of neutrophil functions, contributing to macrophage-polarisation, and inducing T‐cell differentiation. FXII signalling is also involved in activating pro‐inflammatory pathways that induce chemotaxis of leukocytes and increases the vascular- permeability.
The expression of plasminogen activator inhibitor‐1 (PAI‐1), an important factor in the coagulation -process, is induced by TGF‐β. Elevated PAI‐1 has been observed in myocardial -infarction and in cardiac- failure, and the latter is one of the death causes for COVID‐19 patients. Ackermann et al. showed that the expression of the gene for PAI‐1, SERPINE1, is up-regulated in COVID‐19 patients. PAI‐1 is a serine‐protease inhibitor and the main- inhibitor of the tissue‐type (tPA) and the urinary‐type plasminogen activator (uPA). Both are able to activate the plasminogen and, hence, fibrinolysis. Besides the elevated- levels of fibrin in COVID‐19 patients and their resulting blood -clots, increased levels of the fibrin‐degradation product D‐dimer have been detected. A reduced fibrinolysis can cause thrombosis, pulmonary embolus, and it increases the risk of stroke and heart- attack. A case- series was performed to treat blood clots with tPA in COVID‐19 patients. This treatment only showed temporary- improvements [5] (Figure 20).
Figure 20:
SARS‐CoV‐2 S‐protein with a focus on the receptor‐binding domain. The sequences of the spike- protein of human SARS‐CoV‐2, human SARS‐CoV‐1 (75% similarity) and bat RaTG13 (96% similarity) were aligned using MAFFT. The receptor‐binding domain and the ACE2- receptor‐binding region are coloured in light blue. The RGD motif of SARS‐CoV‐2 is high-lighted in red. Numbers refer to the SARS‐CoV‐2 spike protein- sequence. FP, fusion peptide; RGD, amino- acids arginine‐glycine‐aspartic acid; S1, subunit of spike; S2, subun-it of spike (fusion domain); S2´, cleavage site; TM, trans-membrane -domain (Figure 21).
Figure 21: Biological and Clinical Consequences of Integrin Binding via a Rogue RGD Motif in the SARS CoV-2 Spike Protein by Lee Makowski 1,2,William Olson-Sidford 1 and John W. Weisel.
Eun Jeong Park, et al: The spike- glycoprotein attached to the envelope of the SARS-CoV-2 binds to and exploits angiotensin-converting enzyme 2 (ACE2) as an entry receptor to infect pulmonary epithelial- cells. A subset of integrins that recognize the arginyl–glycyl–aspartic acid (RGD) sequence in the cognate- ligands has been predicted in silico to bind the spike -glycoprotein and, thereby, to be exploited for viral- infection. We show experimental- evidence that the β1 integrins predominantly expressed on human pulmonary epithelial -cell lines and primary mouse alveolar epithelial- cells bind to this spike- protein. The cellular β1 integrins support adhesive interactions with the spike- protein independently of ACE2, suggesting the possibility that the β1 integrins may function as an alternative receptor for SARS-CoV-2, which could be targeted for the prevention of viral –infections [6].
Yang, et al: The fact that our candidate RBD- vaccine can induce effective virus-neutralizing activity in three animal species is very encouraging. It is also encouraging to observe (1) that the antibodies shared common binding- epitopes with patients with COVID-19; (2) that there was strong virus-neutralizing activity associated with a good humoral- response; (3) that a simple vaccine- adjuvant such as aluminium could further enhance immune- response; (4) that even a single dose of the vaccine could already generate a high level of virus-neutralizing activity; (5) that the vaccine could protect non-human primates against a challenge with live SARS-CoV-2; and (6) that toxicology -studies in the non-human primates showed that this vaccine candidate is safe to use. our vaccine strategy based on the RBD has been supported by recent- findings that most of the neutralizing- activity is directed against the RBD in the sera of patients with COVID-19 and by evidence from human- monoclonal antibodie [7].
Prompetchara E, et al: An m-RNA vaccine developed by Moderna, expressing whole spike with 2-proline mutations (mRNA-1273) to stabilize the spike protein in prefusion-form, showed a robust Nab- response and protection after viral- challenge in mouse strains and non-human primates. CD4+ T cell and T follicular helper- cell responses were also detected. Wild-type version of spike protein used in viral vector -vaccines, such as ChAdOx1 nCoV-19, developed by the University of Oxford and AstraZeneca was also tested. Immunized -macaques developed NAb and Th1 responses and were mostly protected from viral- replication after challenged [30]. Other spike versions such as RBD trimerization domain using mRNA platform (BNT162b1) developed by BioNTech-Pfizer was also evaluated. Recently, Pfizer reported a direct comparison between BNT162b1 with BNT162b2 which encodes a full-length spike- protein with di-proline mutations. Results showed comparable antibody titers but BNT162b2 provided a more favourable safety-profile. Although the NAb -titers were reported in all vaccine candidates, the results are difficult to compare due to different techniques and readouts in Nab- measurement [8].
Lanying Du, et al: “Angiotensin-converting enzyme 2 ACE 2 has been identified as the receptor of SARS-CoV. A fragment that is located in the S1 subunit and spans amino acids 318–510 is the minimal receptor-binding domain (RBD). Crystallo-graphic- studies have shown the structure of RBD complexed with its receptor ACE2. During the interaction of RBD with the receptor, RBD presents a concave- surface for the N terminus of the receptor peptidase, on which amino- acids 445–460 anchor the entire receptor-binding loop of the RBD -core. This loop (amino acids 424–494 of the RBD), which makes complete contact with the receptor- ACE2, was referred to as receptor-binding motif (RBM). The RBM region is tyrosine rich. Among the 14 residues of RBM that are in direct contact with ACE2, six are tyrosine, representing both the hydroxyl- group and hydrophobic ring. The RBD- region also contains multiple cysteine -residues that are linked by di-sulphidebonds. 2 residues in particular, those at positions 479 and 487, determine SARS disease progression and SARS-CoV tropism (host range). Any residue changes in these 2 positions might therefore enhance animal-to-human or human-to-human transmission [9].
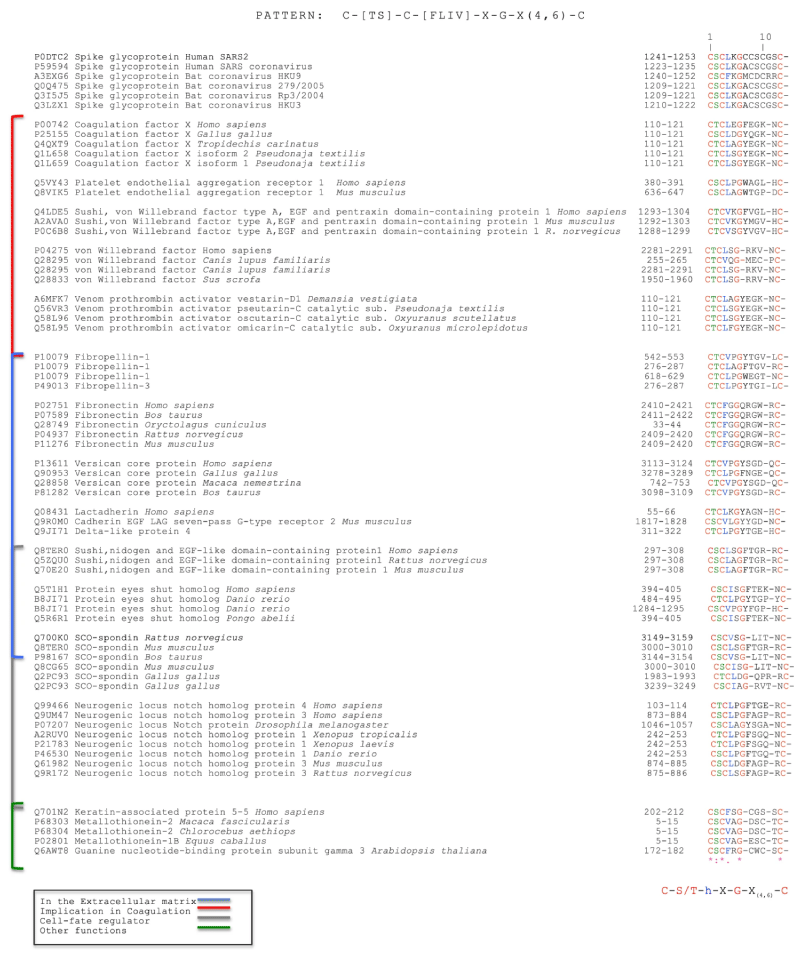
Silvia Buonvino, et al: We analyzed the CD -sequence of CoV-2 S protein in order to identify potential consensus sequence patterns. A bio-informatic analysis using PattInProt v5.4up (https://npsa-prabi.ibcp.fr/cgi-bin/npsa_automat.pl? page=npsa_pattinprot.html)29, setting 100% of similarity, was performed identifying, in 79 proteins out of 561,000 proteins of the UniProt bank, a new potential amino acid AA pattern. The consensus- pattern was C-[TS]-C-h-X-G-X(4,6)-C, herein called CAF-motif (Cysteine Aggregation Fusion), where h is a hydrophobic- residue and X any other residue.
Interestingly, the only viral- proteins that showed the CAF-motif were S -proteins of human corona-viruses SARS and SARS -2 and the S proteins of bat coronavirus. Other proteins involved in coagulation, extracellular recognition and cell- fate presented the same pattern. Intriguingly, the CAF-motif occurs in proteins such as coagulation- factor X, von- Willebrand factor, platelet endothelial- aggregation receptor 1 and some pro-thrombin activators venom toxins that are involved in the coagulation- process. The identification of a common pattern could suggest a new function of the S- protein in the pathological effects of the SARS -2 infection [10] (Figure 22).
Figure 22: Selected Protein Sequences with their UniProt code that show the CAF-motif. From: New Consensus pattern in Spike CoV-2: potential implications in coagulation process and cell–cell fusion.
Sandhya Bangaru, et al: We performed cryo–election microscopy and site-specific glycan analysis of 1 of the leading subunit vaccine- candidates from Novavax, which is based on a full-length spike- protein formulated in polysorbate- 80 detergent. Our studies reveal a stable- prefusion conformation of the spike immunogen with slight differences in the S1 subunit compared with published spike ectodomain structures. We observed interactions between spike- trimers, allowing formation of higher-order spike complexes. This study confirms the structural- integrity of the full-length spike protein immunogen and provides a basis for interpreting immune -responses to this multivalent nano-particle immunogen Glycans on viral glycol-proteins play a wide role in protein folding, stability, and immune recognition and also in facilitating immune evasion.
Analysis of safety and immune-genicity of the Novavax SARS-CoV-2-3Q-2P-FL immunogen in mice and in baboons revealed strong B and T cell- responses to the vaccine with no evidence of vaccine-associated enhanced respiratory-disease [11].
Dominic Wichmann, et al: Median patient age was 73 years (range, 52 to 87 years), 75% of patients were male, and death occurred in the hospital (n = 10) or outpatient sector (n = 2). Coronary heart -disease and asthma or chronic obstructive pulmonary- disease were the most common comorbid conditions (50% and 25%, respectively). Autopsy revealed deep- venous thrombosis in 7 of 12 patients (58%) in whom venous thrombo-embolism was not suspected before death; pulmonary- embolism was the direct cause of death in 4 patients. Postmortem computed tomography revealed reticular infiltration of the lungs with severe bilateral, dense -consolidation, whereas histo-morphologically diffuse alveolar damage was seen in 8 patients. In all patients, SARS-CoV-2 RNA was detected in the lung at high -concentrations; viremia in 6 of 10 and 5 of 12 patients demonstrated high viral RNA- titers in the liver, kidney, or heart [12].
Laura C Price, et al: Huertas, et al. describe the early- pathogenesis in COVID-19 pneumonia defined by a widespread endothelitis affecting multiple organ- systems. As such, viral inclusion bodies are observed within endothelial-cells accompanied by apoptosis, inflammatory cell infiltration and microvascular-thrombosis. At the same time, systemic- inflammation is commonly observed, with elevated levels of C-reactive protein, fibrinogen and cytokines such as interleukin-6. Although the exact mechanisms of COVID-19-induced thrombosis have not been elucidated, at least some of the well-described mechanisms associated with infection/inflammation are likely to be relevant. These include the increased production of tissue -factor and amplification of the coagulation -cascade, resulting in increased production of thrombin and, consequently, fibrin [13].
Simran Preet Kaur, et al: Viral Vectored vaccines, A vaccine- based on viral- vectors is a promising prophylactic solution against a pathogen. These vaccines are highly specific in delivering the genes to the target- cells, highly efficient in the gene transduction, and efficiently induce the immune-response. They offer a long term and high-level of antigenic protein expression and therefore, have a great potential for prophylactic use as these vaccines trigger and prime the cytotoxic T-cells (CTL) which ultimately leads to the elimination of the virus infected- cells [14].
Young Hun Chung, et al: The m-RNA utilized by BioNTech/Pfizer encodes for the RBD Named BNT162b1, the mRNA is modified with single nucleoside incorporations of 1-methylpseudouridine, which not only reduces the immunogenicity of the mRNA in vivo but also increases its translation. The exact mechanism for increased translation has not been entirely elucidated, but 1 hypothesis is that the nucleoside- modification improves RNA stability by decreasing rates of hydrolysis by phospho-diesterases. Instead of m-RNA, viral- vectors use DNA to encode the antigen of interest. Viral vector-vaccines can impart high gene-transduction capabilities due to their ability to enter into cells using the virus’ own receptor for infection, and efficient intracellular trafficking enables high production of target gene- expression.
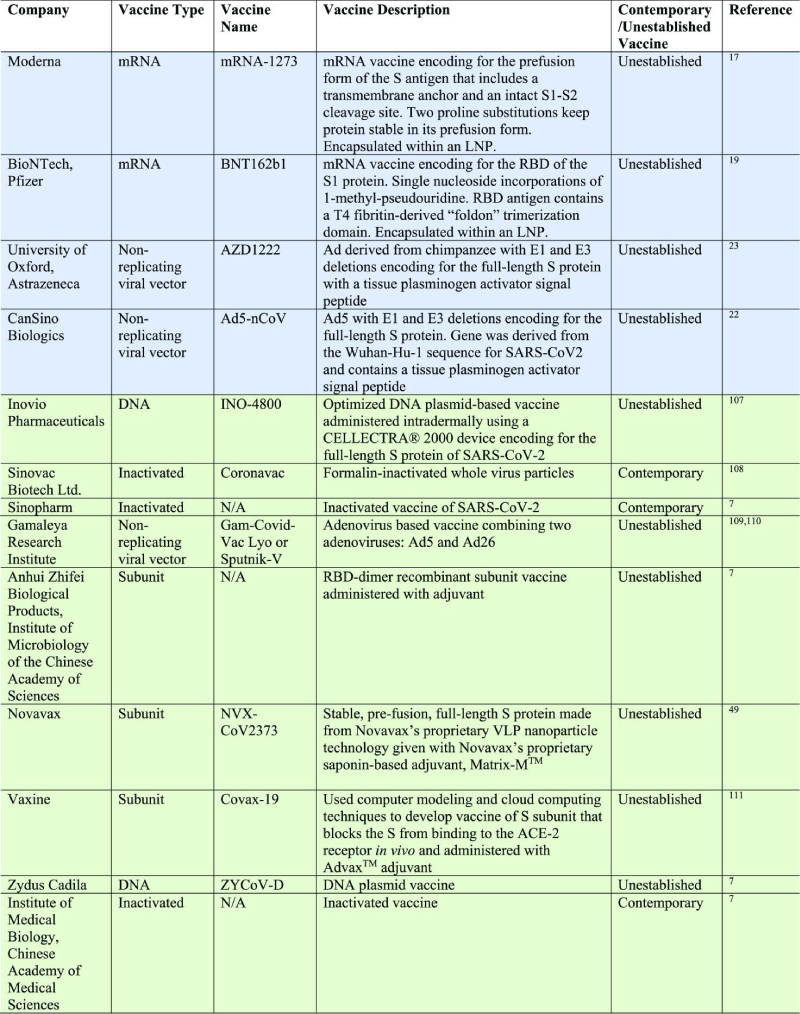
m-RNA vaccines may produce greater amounts of antigen from smaller -doses, but a caveat is that DNA tends to be more stable than m-RNA meaning m-RNA expression is generally shorter lived. Oxford/Astra zenca. Ads are common cold causing viruses that have a double-stranded DNA genome. Specifically, CanSino is utilizing Ad type 5 (Ad5), giving the vaccine the name Ad5-nCoV.21 Oxford/Astra-zeneca is employing a different viral- vector, an Ad derived from the chimpanzee (the use of the chimpanzee- vector minimizes possible interaction with prevalent antibodies against Ads), which was subsequently named AZD1222.23 Ad5-nCoV specifically encodes for the full-length S protein of SARS-CoV-2, unlike both Moderna and BioNTech/Pfizer, which both encoded subunits of the S -protein. The gene was derived from the Wuhan-Hu-1 sequence for SARS-CoV-2 and, along with a tissue- plasminogen activator signal peptide, was cloned into an E1 and E3 deleted Ad5 vector. Deletion of E1 inactivates the replication potential of the vaccine while deletion of E3 allows for the insertion of larger- genes up to 8 kb.113 [15] (Table 1).
Table 1: Descriptions of Vaccines That Have Moved Beyond Their Initial Safety and Immunogenicity Phase I Study.
Experimental projecty hypotheys
In order to verify the procoagulant activities of spike protein it is interesting to test different kind of Situation:
Full lenght spike protein
Subunity
RBD domain
Mutated spike (like in some m RNA vaccine).
All this molecules must to be added to human plasma and blood in in vitro condition and at various concentration. (To simulate a COVID lung severe disease with clots- HIGH SPIKE CONCENTRATION, or a vaccination procedure low SPIKE production).
After a right time and temperature it must be verified coagulation activation level or not and platelet activation and aggregation.
This kind of experiment make possible to verify the single contribute of SPIKE full length or its subunits, or by its Domains in the coagulative process.
By literature and related nowadays level of COVID -19 development various strategy have been and are used to produce COVID-19 vaccine.
Because the various pharmaceutical company chosen different strategy it is interesting to verify the contribute of the SPIKE protein or its subunits and domains in a rare effect recognized as some central thrombosis.
A real fact that happened in this days is that some vaccine producers updated their technical schede adding this rare event as side effect ADR. (And by regulative institution verify).
For this reason the scope of this work is to verify if COVID-19 coagulopathy is or not related to the vaccine rare effect showed. (Physiopathology similar?)
Obviously the concentration-level of SPIKE protein during a severe situation of pulmonitis and its coagulopathy is a high level of noxa versus the spike production by the vaccine shot and after the viral genetic transduction phases.
But it is interesting to verify that the clotting factors (in other classic physio-pathological setting) are activated following kinetics that amplify the signal.
Related the literature reported and to the rare side effect ADR of some covid- 19 vaccine like thrombosis it is needed to search new strategy in vaccine design.
The next generation of COVID-19 vaccine must to take in consideration the role played by RGD domain, or SUBUNITS or FULL LENGTS spike, or by MUTATED SPIKE to link some integrin like fibronectin or to activate other factor involved in clotting cascade.
The same the role played by the medium in the glycosylation pattern must to be deeply investigated.
And finally: Effect like amplification of signal and level of gene transduction play a role for the various vaccines rare thrombosys effect?
The role played by the molecule cited in this article it is clear by science.
Ethical consideration: all international rules applied.
- Shulla A, Gallagher T. Role of Spike Protein Endodomains in Regulating Coronavirus Entry. Protein Struct Fold. 2009; 284: 32725-32734. PubMed: https://pubmed.ncbi.nlm.nih.gov/19801669/
- Grobbelaar LM, Venter C, Vlok M, Ngoepe M, Laubscher GJ, Lourens PJ, et al. SARS-CoV-2 spike protein S1 induces fibrin(ogen) resistant to fibrinolysis: Implications for microclot formation in COVID-19.
- Janardhan V, Janardhan V, Kalousek V. COVID-19 as a Blood Clotting Disorder Masquerading as a Respiratory Illness: A Cerebrovascular Perspective and Therapeutic Implications for Stroke Thrombectomy. J Neuroimaging. 2020; 30: 555-561. PubMed: https://pubmed.ncbi.nlm.nih.gov/32776617/
- Makowski L, Olson-Sidford W, Weisel J. Biological and Clinical Consequencesof Integrin Binding via a Rogue RGDMotif in the SARS CoV-2 Spike. Protein. Viruses. 2021; 13: 146. PubMed: https://pubmed.ncbi.nlm.nih.gov/33498225/
- Carvacho I, Piesche M. RGD‐binding integrins and TGF‐β in SARS‐CoV‐2 infections – novel targets to treat COVID‐19 patients? Clin Transl Immunol. 2021; 10: e1240. PubMed: https://pubmed.ncbi.nlm.nih.gov/33747508/
- Park EJ, Myint PK, Appiah MG, Darkwah S, Caidengbate S, et al. The Spike Glycoprotein of SARS-CoV-2 Binds to β1 Integrins Expressed on the Surface of Lung Epithelial Cells. Viruses. 2021; 13: 645. PubMed: https://pubmed.ncbi.nlm.nih.gov/33918599/
- Yang J, Wang W, Chen Z, Lu S, Yang F, et al. A vaccine targeting the RBD of the S protein of SARS-CoV-2 induces protective immunity. Nature. 2020; 586: 572–577. PubMed: https://pubmed.ncbi.nlm.nih.gov/32726802/
- Prompetchara E, Ketloy C, Tharakhet K, Kaewpang P, Buranapraditkun S, et al. DNA vaccine candidate encoding SARS-CoV-2 spike proteins elicited potent humoral and Th1 cell-mediated immune responses in mice. PLoS One 2021; 16: e0248007. PubMed: https://pubmed.ncbi.nlm.nih.gov/33750975/
- Du L, He Y, Zhou Y, Liu S, Zheng BJ, et al. The spike protein of SARS-CoV — a target for vaccine and therapeutic development. Nat Rev Microbiol. 2009; 7: 226–236. PubMed: https://pubmed.ncbi.nlm.nih.gov/19198616/
- Buonvino S, Melino S. New Consensus pattern in Spike CoV-2: potential implications in coagulation process and cell–cell fusion. Cell Death Discovery. 2020; 6: 134.
- Bangaru S, Ozorowski G, Turner HL, Antanasijevic A, Huang D, et al. Structural analysis of full-length SARS-CoV-2 spike protein from an advanced vaccine candidate. Science. 2020; 370: 1089-1094. PubMed: https://pubmed.ncbi.nlm.nih.gov/33082295/
- Wichmann D, Sperhake JP, Lütgehetmann M, Steurer S, Edler C, et al. Autopsy Findings and Venous Thromboembolism in Patients With COVID-19: A Prospective Cohort Study. Ann Intern Med. 2020; 173: 268-277. PubMed: https://pubmed.ncbi.nlm.nih.gov/32374815/
- Price LC, McCabe C, Garfield B, Wort SJ. Thrombosis and COVID-19 pneumonia: the clotthickens! Eur Respir J. 2020; 56: 2001608. PubMed: https://pubmed.ncbi.nlm.nih.gov/32554532/
- Kaur SP, Gupta V. COVID-19 Vaccine: A comprehensive status report. Virus Res. 2020; 288: 198114. PubMed: https://pubmed.ncbi.nlm.nih.gov/32800805/
- Chung YH, Beiss V, Fiering SN, Steinmetz NF. COVID-19 Vaccine Frontrunners and Their Nanotechnology Design. ACS Nano. 2020; 14: 12522-12537. PubMed: https://pubmed.ncbi.nlm.nih.gov/33034449/
- Duan L, Zheng Q, Zhang H, Niu Y, Lou Y, et al. The SARS-CoV-2 Spike Glycoprotein Biosynthesis, Structure, Function, and Antigenicity: Implications for the Design of Spike-Based Vaccine Immunogens ront Immunol. 2020; 11: 576622. PubMed: https://pubmed.ncbi.nlm.nih.gov/33117378/
- Xia X, Lundstrom K, Alaa AA. Aljabali. Domains and Functions of Spike Protein in SARS-Cov-2 in the Context of Vaccine Design. Viruses. 2021; 13: 109. PubMed: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7829931/
- Kamel MH, Yin W, Zavaro C, Francis JM, Chitalia VC. Cells Review Hyperthrombotic Milieu in COVID-19 Patients. Cells. 2020 31; 9: 2392. PubMed: https://pubmed.ncbi.nlm.nih.gov/33142844/
- Malik JA, Mulla AH, Farooqi T, Pottoo FH, Anwar S. Targets and strategies for vaccine development against SARS-CoV-2. Biomed Pharmacother. 2021; 137: 111254. PubMed: https://pubmed.ncbi.nlm.nih.gov/33550049/
- Dai L, Gao GF. Viral targets for vaccines against COVID-19. Nat Rev Immunol. 2021; 21: 73–82. PubMed: https://pubmed.ncbi.nlm.nih.gov/33340022/
- Zaid Y, Puhm F, Allaeys I, Naya A, Oudghiri M, et al. Platelets Can Associate With SARS-CoV-2 RNA and Are Hyperactivated in COVID-19. Circ Res. 2020; 127: 1404–1418. PubMed: https://pubmed.ncbi.nlm.nih.gov/32938299/
- Beddingfield B, Iwanaga N, Zheng W, Roy CJ, Hu TY, et al. The Integrin Binding Peptide, ATN-161, as a Novel Therapy for SARS-CoV-2 Infection. JACC Basic Transl Sci. 2021; 6: 1-8. PubMed: https://pubmed.ncbi.nlm.nih.gov/33102950/
- Mészáros B, Sámano-Sánchez H, Alvarado-Valverde J, Čalyševa J, Martínez-Pérez E, et al. Short linear motif candidates in the cell entry system used by SARS-CoV-2 and their potential therapeutic implications. Sci Signal. 2021; 14: eabd0334. PubMed: https://pubmed.ncbi.nlm.nih.gov/33436497/
- Zana X, Sitasuwana P, Powellb J, Dreherb TW, Wanga Q. Polyvalent display of RGD motifs on turnip yellow mosaic virus for enhanced stem cell adhesion and spreading. Acta Biomater. 2012; 8: 2978-2985. PubMed: https://pubmed.ncbi.nlm.nih.gov/22522012/
- Wang Y, Carrim N, Ni H. Fibronectin orchestrates thrombosis and hemostasis. Oncotarget. 2015; 6: 19350–19351. PubMed: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4637283/
- Vadasz B, Chen P, Yougbaré I, Zdravic D, Li J, et al. Article Feb 2015 Platelets and platelet alloantigens: Lessons from human patients and animal models of fetal and neonatal alloimmune thrombocytopenia. Genes Dis. 2015; 2: 173-185. PubMed: https://pubmed.ncbi.nlm.nih.gov/28345015/